Methods and Analyses
MIDAA
Multiomics Integration via Deep Archetypal Analysis (MIDAA)
MIDAA is a package designed for performing Deep Archetypal Analysis on multiomics data.
Reference: Milite 2025, Genome Biology
Code: https://github.com/sottorivalab/midaa
NEUROVELO
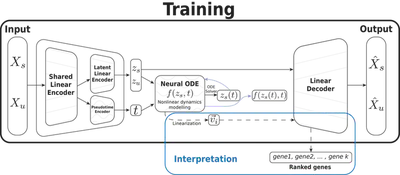
NeuroVelo: interpretable learning of cellular dynamics
NeuroVelo: physics-based interpretable learning of cellular dynamics. It is implemented on Python3 and PyTorch, the model estimate velocity field and genes that drives the splicing dynamics.
Reference: Kouadri Boudjelthia, 2024, biorxiv
https://github.com/idriskb/NeuroVelo
EPICC ANALYSIS
EPICC papers data analysis pipelines
EPICC analysis (Heide, Househam, et al. 2022; Househam, Heide et al. 2022)
https://github.com/sottorivalab/EPICC2021_data_analysis_EPIGENOME
https://github.com/sottorivalab/EPICC2021_data_analysis_RNA
EPICC SIMULATIONS AND INFERENCE
EPICC simulations and inference
EPICC second paper simulation and inference
Reference: Househam, Heide et al. 2022
https://github.com/sottorivalab/EPICC2021_inference
https://github.com/T-Heide/MLLPT
MOBSTER
Subclonal reconstruction in cancer by combining evolutionary theory with machine learning
Reference: Caravagna et al. 2020
Code: https://github.com/caravagnalab/mobster
MCMC - MUTATIONAL DISTANCES
Inference of de-coupled single cell microscopic parameters such as the mutation rates per division and the cell death rates from multi-sampling genomic data
Reference: Werner et al. 2020
Code: https://github.com/sottorivalab/MCMC-MutationalDistances-
CHESS
A spatial model of tumour growth that also simulates different sampling strategies and the generation of genomic data:
Reference: CHESS Chkhaidze et al. 2019
Code: https://github.com/sottorivalab/CHESS.cpp
REVOLVER
Detecting repeated evolutionary trajectories in cancer using Transfer Learning:
Reference: Caravagna et al. 2018
Code: https://github.com/sottorivalab/revolver
QUANTIFYING-SELECTION
A set of tools to measure neutral evolution and subclonal selection using either a frequentist approach or an Approximate Bayesian Computation approach for model selection
Reference: Quantifying-Selection (Williams et al. 2016; Williams et al. 2018)
Code: https://marcjwilliams1.github.io/quantifying-selection