Develop tools that combine AI with mechanistic modelling for oncology
Identify the biological rules that allow cells to build tissues in cancer and normal
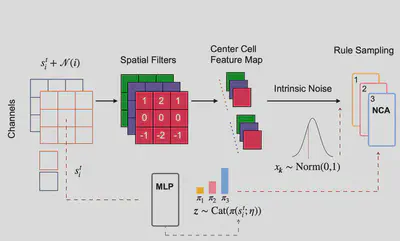
Neural Cellular Automata (NCA) are a Deep Learning version of Cellular Automata (CA) that leverage back-propagation to automatically identify CA rules from the data. We designed a new AI architecture termed Mixture of Neural Cellular Automata to handle biological stochasticity in NCAs, and allow their use to identify the spatial rules of tissue formation leveraging on time-series spatial biology data Milite 2025, biorxiv.
Quantify cellular dynamics from singe cell multi-omics
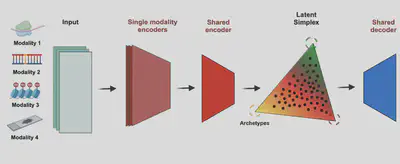
Single cell technologies have revolutionised biomedical research, and new machine learning methods allow reducing the dimensionality of sparse and noisy data to tractable latent spaces. However, the results of those methods remain hard to interpret due to the inherent non-linearity in their transformations, and the lack of interpretability in terms of biological mechanisms.
We developed two orthogonal methods to make sense to single cell data in light of evolution and dynamical systems theory. First, we leveraged on Archetypes Analysis to measure the evolutionary trade-offs between cellular programmes with MIDAA Milite 2025, Genome Biology. Second, in collaboration with the Sanguinetti Lab at SISSA (Triest) we used Neural Ordinary Differential Equations to reconstruct gene regulatory networks that drive the temporal dynamics of single cell data with a new tool called NeuroVelo Kouadri Boudjelthia 2024, biorxiv.
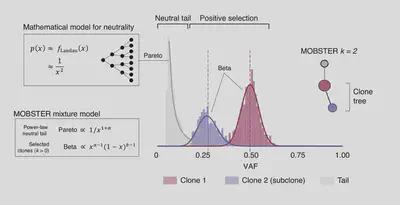
Measure clonal evolution with MOBSTER
We present a novel approach for model-based tumor subclonal reconstruction, called MOBSTER, which combines machine learning with theoretical population genetics. Using public whole-genome sequencing data from 2,606 samples from different cohorts, new data and synthetic validation, we show that this method is more robust and accurate than current techniques (Caravagna et al. 2020, Nature Genetics).
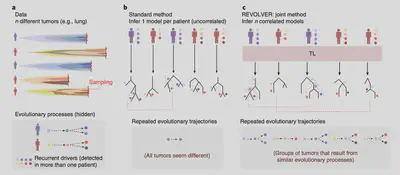
Detecting repeated cancer evolution in large genomic datasets
We developed REVOLVER (Repeated EVOLution in cancER), a tool based on a type of machine learning approach called ‘Transfer Learning’, to analyse and compare sets of phylogenetic trees from multi-region sequencing studies in cancer, with the aim of identifying hidden patterns of repeated evolutionary trajectories in the data (Caravagna et al. 2018, Nature Methods). We analysed a total of 768 samples from 178 patients with lung, colon, breast and renal cancer, and found that some of these repeated evolutionary trajectories also correlated with prognosis.