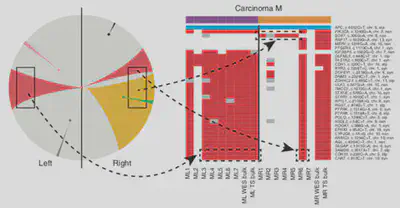
Measure and predict cancer evolution
Measuring the co-evolution of cancer and the patient’s immune system
We quantified how cancer neoantigens, mutations that stimulate the response of the immune system, evolve during tumourigenesis and how evolutionary forces such as immune-mediated negative selection (Lakatos 2020, Nature Genetics; Zapata 2023, Nature Genetics) or epigenetic rewiring (Lakatos 2025, Nature Genetics) change the repertoire of tumour immunogenic variants.
Darwinian genetic and epigenetic evolution vs non-Darwinian cell plasticity in colorectal cancer
We concomitantly measured multiple layers of cellular information in colorectal cancers. We uncovered novel epigenetic drivers and tumourigenic processes that occurred without genetic alterations but instead through re-wiring of the chromatin (Heide, Househam et al. 2022, Nature). We also found that a significant proportion of transcriptional variation within cancers results from cell phenotypic plasticity, with no direct genetic control (Househam, Heide et al. 2022, Nature).
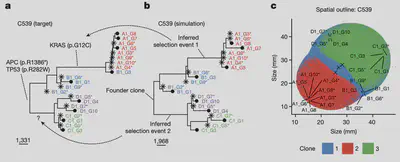
Predicting recurrence in prostate cancer
We found that evolutionary divergence at the genetic level but also at the cell morphology level (Gleason grade), was a strong independent predictor of 10y recurrence in advanced localized prostate cancer (Fernandez-Mateos et al. 2024, Nature Cancer).
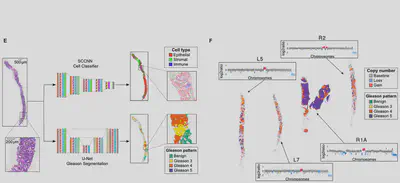
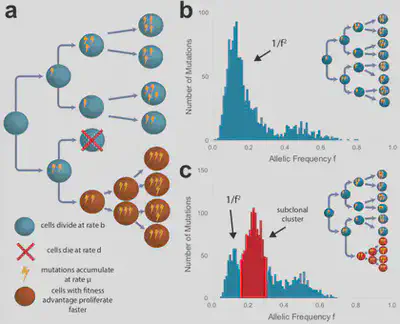
Measuring neutral evolution and positive selection from patient genomic data
Inspired by the Big Bang study, we used genetic data of more than 900 tumours from 14 different types to understand the patterns of tumour expansion in large cohorts. We found that many cancer mutational patterns, despite appearing complex, could be explained by a simple mathematical model of neutral tumour evolution (Williams, Werner et al. 2016, Nature Genetics). We then quantified also non-neutral evolutionary dynamics in those cancers where subclonal selection was detectable (Williams et al. 2018, Nature Genetics).
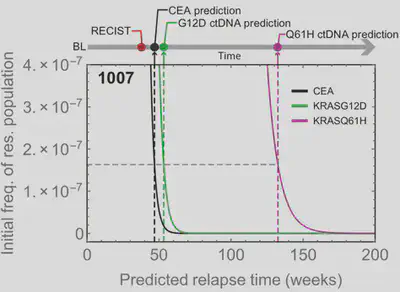
Mathematical modelling of treatment resistance applied to clinical trials
We combined frequent circulating tumour DNA profiling over time (every 4 weeks) of colorectal cancer patients undergoing EGFR targeted treatment within a phase II trial, with mathematical modelling of treatment resistance for quantitative forecasting of the waiting time to progression (Khan, Cunningham, Werner et al. 2018, Cancer Discovery).
‘Big Bang’ tumour growth
We demonstrated that, after having accumulated a ‘jackpot’ set of driver alterations, many colorectal cancers grow as a single “Big Bang” expansion, populated by numerous intermixed subclones that are not subject to stringent selection (Sottoriva et al. 2015, Nature Genetics). Additional evidence of lack of subclonal selection in established colorectal cancers has been presented in subsequent collaborative work (Sun et al. 2017, Nature Genetics; Cross et al. 2018, Nature Ecology and Evolution; Househam, Heide et al. 2022, Nature).